About
dsCheck especially focuses on off-target searching for 'long' dsRNA-mediated RNAi, which is widely used to knockdown gene expression in non-mammalian species including Drosophila, C. elegans, Arabidopsis, and so on. The software 'dices' the input sequence into 19-nt siRNA coctails, and performs exhaustive scan for each 'diced siRNA' to find off-target gene candidates, simulating the biochemical process of dsRNA-mediated RNAi in vivo.
This website provides ...
- Off-target verification of your dsRNA sequences.
[link]
- Optimal design of off-target minimized dsRNA, when off-target effects are suspected.
[link]
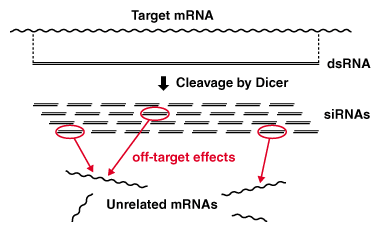
Biochemical process of dsRNA-mediated RNAi. Off-target effects are caused by 'diced' siRNAs that have sequence similarities with unrelated genes.
Examples
Verify your dsRNA [link]
- dsCheck accepts FASTA format or flat nucleotide sequence as an input. Users can also enter an accession number to retrieve a sequence from GenBank. In the input sequence, characters other than A, T, G, C, U, N, R, Y, M, K, S, W, H, B, V and D are ignored. U is replaced with T, and letters other than A, T, G, C and U are replaced with Ns. Both lower-case and upper-case letters are accepted. Current limit of the input sequence is 3000 bp.
- Choose your species. Currently, RefSeq mRNA sequences for Drosophila, C. elegans, Arabidopsis and Oryza sativa are available.
- Click 'verify' to show off-target gene candidates.
- Suspected off-target gene candidates are listed in few seconds. Number of hits with a complete match (19/19 matches), one mismatches (18/19 matches), or two mismatches (17/19) are counted for every off-target gene candidates individually. In this example, significant hits against two splicing variants of pdm2 (maybe your intended target gene), and two other genes nub and vvl (these seem to be off-targets). This result indicates a high risk of cross-suppressing nub and vvl by your dsRNA. Intended target genes (NM_078834.2, pdm2 and , NM_165017.1pdm2) are highlighted.
- The result can be downloaded as a 'Tab-Text' file.
Design off-target minimized dsRNA [link]
- dsCheck accepts FASTA format or flat nucleotide sequence as an input. Users can also enter an accession number to retrieve a sequence from GenBank. In the input sequence, characters other than A, T, G, C, U, N, R, Y, M, K, S, W, H, B, V and D are ignored. U is replaced with T, and letters other than A, T, G, C and U are replaced with Ns. Both lower-case and upper-case letters are accepted. Current limit of the input sequence is 3000 bp.
- Choose your species. Currently, RefSeq mRNA sequences for Drosophila, C. elegans, Arabidopsis and Oryza sativa are available.
- Select dsRNA length to design.
- Click 'design' to design off-target minimized dsRNA for your input sequence.
- Giving the condition that the dsRNA length is 100 bp, the server returns off-target minimized region, 1741-1840. This 100 bp dsRNA has only 4 hits with 17/19 matches against the nearest off-target gene.
- These sequences (NM_165017.1, pdm2 and NM_078834.2, pdm2) are regarded as the intended target genes.
References
- Naito,Y.*, Yamada,T.*, Matsumiya,T., Ui-Tei,K., Saigo,K. and Morishita,S. (2005) dsCheck: highly sensitive off-target search software for dsRNA-mediated RNA interference. Nucleic Acids Res., in press. * Joint First Authors.
- Yamada,T. and Morishita,S. (2005) Accelerated off-target search algorithm for siRNA. Bioinformatics, 21, 1316-1324.
- Naito,Y.*, Yamada,T.*, Ui-Tei,K., Morishita,S. and Saigo,K. (2004) siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res., 32, W124-W129. * Joint First Authors.
- Ui-Tei,K., Naito,Y., Takahashi,F., Haraguchi,T., Ohki-Hamazaki,H., Juni,A., Ueda,R. and Saigo,K. (2004) Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res., 32, 936-948.
Credits
Yuki Naito, Tomoyuki Yamada, Takahiro Matsumiya, Kumiko Ui-Tei, Shinichi Morishita, and Kaoru Saigo used the expertise of biology and informatics, and designed the system.
Tomoyuki Yamada and Takahiro Matsumiya implemented the entire system of dsCheck, and developed novel, efficient algorithms for enumerating potential off-target candidates. Yuki Naito and Shinichi Morishita together assisted Tomoyuki Yamada in improving several prototypes of dsCheck.
This work was supported in part by the Special Coordination Fund for Promoting Science and Technology to Kaoru Saigo, the Leading Project for Biosimulation to Shinichi Morishita, and Grants-in-Aid for Scientific Research to Kumiko Ui-Tei, Kaoru Saigo and Shinichi Morishita from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
|
 Instructions
on how to use dsCheck
Instructions
on how to use dsCheck
 Instructions
on how to use dsCheck
Instructions
on how to use dsCheck